QuSoft & CWI, Amsterdam, the Netherlands.
Find this paper interesting or want to discuss? Scite or leave a comment on SciRate.
Abstract
We present three quantum algorithms for clustering graphs based on higher-order patterns, known as motif clustering. One uses a straightforward application of Grover search, the other two make use of quantum approximate counting, and all of them obtain square-root like speedups over the fastest classical algorithms in various settings. In order to use approximate counting in the context of clustering, we show that for general weighted graphs the performance of spectral clustering is mostly left unchanged by the presence of constant (relative) errors on the edge weights. Finally, we extend the original analysis of motif clustering in order to better understand the role of multiple `anchor nodes’ in motifs and the types of relationships that this method of clustering can and cannot capture.
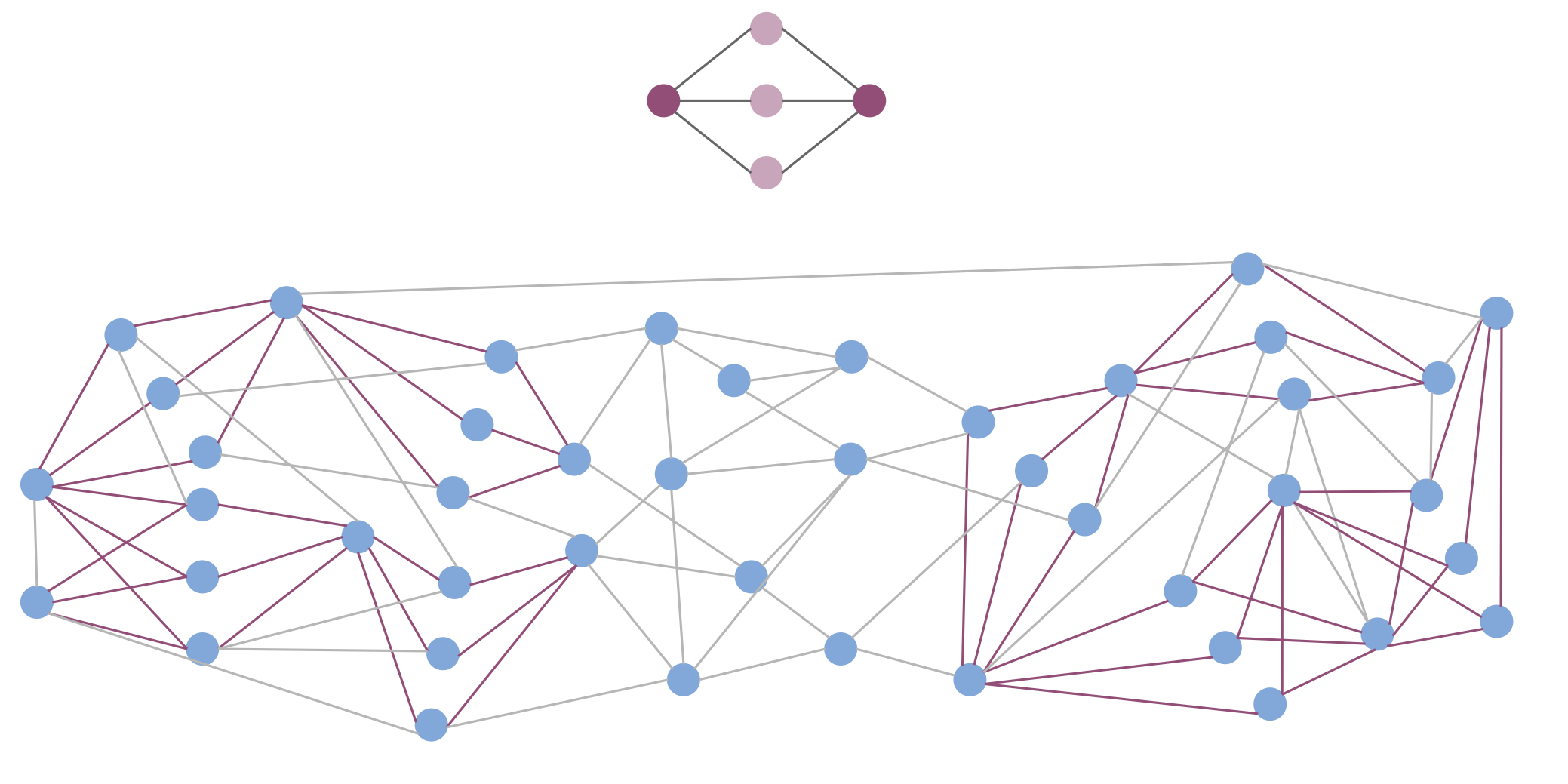
Featured image: An example motif (top), and the connectivity of a graph (bottom) with respect to that motif. Edges highlighted in red demonstrate participation in a motif. Groups of vertices that are connected strongly through motif (red) edges are more likely to be put into the same clusters.
Popular summary
► BibTeX data
► References
[1] Simon Apers and Ronald de Wolf. Quantum speedup for graph sparsification, cut approximation and Laplacian solving. In 2020 IEEE 61st Annual Symposium on Foundations of Computer Science (FOCS), pages 637–648. IEEE, 2020. arXiv:1911.07306. doi:https://doi.org/10.1109/FOCS46700.2020.00065.
https://doi.org/10.1109/FOCS46700.2020.00065
arXiv:1911.07306
[2] Réka Albert, Hawoong Jeong, and Albert-László Barabási. Diameter of the world-wide web. Nature, 401(6749):130–131, 1999. doi:https://doi.org/10.1038/43601.
https://doi.org/10.1038/43601
[3] Reka Albert. Scale-free networks in cell biology. Journal of cell science, 118(21):4947–4957, 2005. doi:https://doi.org/10.1242/jcs.02714.
https://doi.org/10.1242/jcs.02714
[4] Daron Acemoglu, Asuman Ozdaglar, and Alireza Tahbaz-Salehi. Systemic risk and stability in financial networks. American Economic Review, 105(2):564–608, 2015. doi:https://doi.org/10.1257/aer.20130456.
https://doi.org/10.1257/aer.20130456
[5] Albert-László Barabási and Eric Bonabeau. Scale-free networks. Scientific American, 288(5):60–69, 2003. doi:https://doi.org/10.1126/science.1173299.
https://doi.org/10.1126/science.1173299
[6] Michel Boyer, Gilles Brassard, Peter Høyer, and Alain Tapp. Tight bounds on quantum searching. Fortschritte der Physik: Progress of Physics, 46(4-5):493–505, 1998. arXiv:quant-ph/9605034. doi:https://doi.org/10.1002/(SICI)1521-3978(199806)46:4/5<493::AID-PROP493>3.0.CO;2-P.
<a href="https://doi.org/10.1002/(SICI)1521-3978(199806)46:4/53.0.CO;2-P”>https://doi.org/10.1002/(SICI)1521-3978(199806)46:4/5<493::AID-PROP493>3.0.CO;2-P
arXiv:quant-ph/9605034
[7] Tom Britton, Maria Deijfen, and Anders Martin-Löf. Generating simple random graphs with prescribed degree distribution. Journal of statistical physics, 124(6):1377–1397, 2006. doi:https://doi.org/10.1007/s10955-006-9168-x.
https://doi.org/10.1007/s10955-006-9168-x
[8] Austin R. Benson, David F. Gleich, and Jure Leskovec. Higher-order organization of complex networks. Science, 353(6295):163–166, July 2016. arXiv:1612.08447. doi:10.1126/science.aad9029.
https://doi.org/10.1126/science.aad9029
arXiv:1612.08447
[9] Gilles Brassard, Peter Høyer, Michele Mosca, and Alain Tapp. Quantum amplitude amplification and estimation. Contemporary Mathematics, 305:53–74, 2002. arXiv:quant-ph/0005055. doi:https://doi.org/10.1090/conm/305/05215.
https://doi.org/10.1090/conm/305/05215
arXiv:quant-ph/0005055
[10] Béla Bollobás, Svante Janson, and Oliver Riordan. The phase transition in inhomogeneous random graphs. Random Structures & Algorithms, 31(1):3–122, 2007. doi:https://doi.org/10.1002/rsa.20168.
https://doi.org/10.1002/rsa.20168
[11] Ryan Babbush, Jarrod R McClean, Michael Newman, Craig Gidney, Sergio Boixo, and Hartmut Neven. Focus beyond quadratic speedups for error-corrected quantum advantage. PRX Quantum, 2(1):010103, 2021. arXiv:2011.04149. doi:https://doi.org/10.1103/PRXQuantum.2.010103.
https://doi.org/10.1103/PRXQuantum.2.010103
arXiv:2011.04149
[12] Marián Boguná and Romualdo Pastor-Satorras. Class of correlated random networks with hidden variables. Physical Review E, 68(3):036112, 2003. doi:https://doi.org/10.1103/PhysRevE.68.036112.
https://doi.org/10.1103/PhysRevE.68.036112
[13] Danielle S Bassett and Olaf Sporns. Network neuroscience. Nature neuroscience, 20(3):353–364, 2017. doi:https://doi.org/10.1038/nn.4502.
https://doi.org/10.1038/nn.4502
[14] Fan Chung and Linyuan Lu. The average distances in random graphs with given expected degrees. Proceedings of the National Academy of Sciences, 99(25):15879–15882, 2002. doi:https://doi.org/10.1073/pnas.252631999.
https://doi.org/10.1073/pnas.252631999
[15] T-H Hubert Chan, Anand Louis, Zhihao Gavin Tang, and Chenzi Zhang. Spectral properties of hypergraph Laplacian and approximation algorithms. Journal of the ACM (JACM), 65(3):1–48, 2018. arXiv:1605.01483. doi:https://doi.org/10.1145/3178123.
https://doi.org/10.1145/3178123
arXiv:1605.01483
[16] Norishige Chiba and Takao Nishizeki. Arboricity and subgraph listing algorithms. SIAM Journal on computing, 14(1):210–223, 1985. doi:https://doi.org/10.1137/0214017.
https://doi.org/10.1137/0214017
[17] Michalis Faloutsos, Petros Faloutsos, and Christos Faloutsos. On power-law relationships of the internet topology. In The Structure and Dynamics of Networks, pages 195–206. Princeton University Press, 2011. doi:https://doi.org/10.1145/316194.316229.
https://doi.org/10.1145/316194.316229
[18] Prasanna Gai and Sujit Kapadia. Contagion in financial networks. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences, 466(2120):2401–2423, 2010. doi:https://doi.org/10.1098/rspa.2009.0410.
https://doi.org/10.1098/rspa.2009.0410
[19] Ajem Janssen, Johan van Leeuwaarden, and Seva Shneer. Counting cliques and cycles in scale-free inhomogeneous random graphs. Journal of Statistical Physics, 175(1):161–184, 2019. arXiv:1812.04384. doi:https://doi.org/10.1007/s10955-019-02248-w.
https://doi.org/10.1007/s10955-019-02248-w
arXiv:1812.04384
[20] Ioannis Koutis, Alex Levin, and Richard Peng. Faster spectral sparsification and numerical algorithms for SDD matrices. ACM Transactions on Algorithms (TALG), 12(2):1–16, 2015. arXiv:1209.5821. doi:https://doi.org/10.1145/2743021.
https://doi.org/10.1145/2743021
arXiv:1209.5821
[21] Matthieu Latapy. Main-memory triangle computations for very large (sparse (power-law)) graphs. Theoretical computer science, 407(1-3):458–473, 2008. doi:https://doi.org/10.1016/j.tcs.2008.07.017.
https://doi.org/10.1016/j.tcs.2008.07.017
[22] Andrea Lancichinetti, Santo Fortunato, and Filippo Radicchi. Benchmark graphs for testing community detection algorithms. Physical review E, 78(4):046110, 2008. arXiv:0805.4770. doi:https://doi.org/10.1103/PhysRevE.78.046110.
https://doi.org/10.1103/PhysRevE.78.046110
arXiv:0805.4770
[23] Gipsi Lima-Mendez and Jacques van Helden. The powerful law of the power law and other myths in network biology. Molecular BioSystems, 5(12):1482–1493, 2009. doi:https://doi.org/10.1039/B908681A.
https://doi.org/10.1039/B908681A
[24] Ali Masoudi-Nejad, Falk Schreiber, and Zahra Razaghi Moghadam Kashani. Building blocks of biological networks: a review on major network motif discovery algorithms. IET systems biology, 6(5):164–174, 2012. arXiv:1804.06990. doi:https://doi.org/10.1049/iet-syb.2011.0011.
https://doi.org/10.1049/iet-syb.2011.0011
arXiv:1804.06990
[25] Dror Marcus and Yuval Shavitt. Efficient counting of network motifs. In 2010 IEEE 30th International Conference on Distributed Computing Systems Workshops, pages 92–98. IEEE, 2010. doi:https://doi.org/10.1109/ICDCSW.2010.41.
https://doi.org/10.1109/ICDCSW.2010.41
[26] Nataša Pržulj. Biological network comparison using graphlet degree distribution. Bioinformatics, 23(2):e177–e183, 2007. doi:https://doi.org/10.1093/bioinformatics/btl301.
https://doi.org/10.1093/bioinformatics/btl301
[27] Richard Peng, He Sun, and Luca Zanetti. Partitioning well-clustered graphs: Spectral clustering works! In Conference on Learning Theory, pages 1423–1455. PMLR, 2015. arXiv:1411.2021. doi:https://doi.org/10.1137/15M1047209.
https://doi.org/10.1137/15M1047209
arXiv:1411.2021
[28] Olaf Sporns and Rolf Kötter. Motifs in brain networks. PLoS Biol, 2(11):e369, 2004. doi:https://doi.org/10.1371/journal.pbio.0020369.
https://doi.org/10.1371/journal.pbio.0020369
[29] Shai S Shen-Orr, Ron Milo, Shmoolik Mangan, and Uri Alon. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics, 31(1):64–68, 2002. doi:https://doi.org/10.1038/ng881.
https://doi.org/10.1038/ng881
[30] Daniel A Spielman and Shang-Hua Teng. Spectral partitioning works: Planar graphs and finite element meshes. Linear Algebra and its Applications, 421(2-3):284–305, 2007. doi:https://doi.org/10.1109/SFCS.1996.548468.
https://doi.org/10.1109/SFCS.1996.548468
[31] Daniel A Spielman and Shang-Hua Teng. Nearly linear time algorithms for preconditioning and solving symmetric, diagonally dominant linear systems. SIAM Journal on Matrix Analysis and Applications, 35(3):835–885, 2014. arXiv:cs/0607105. doi:https://doi.org/10.1137/090771430.
https://doi.org/10.1137/090771430
arXiv:cs/0607105
[32] Steven H Strogatz. Exploring complex networks. Nature, 410(6825):268–276, 2001. doi:https://doi.org/10.1038/35065725.
https://doi.org/10.1038/35065725
[33] Yuuki Takai, Atsushi Miyauchi, Masahiro Ikeda, and Yuichi Yoshida. Hypergraph clustering based on pagerank. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pages 1970–1978, 2020. arXiv: 2006.08302. doi:https://doi.org/10.1145/3394486.3403248.
https://doi.org/10.1145/3394486.3403248
arXiv:2006.08302
[34] Ulrike von Luxburg. A tutorial on spectral clustering. Statistics and computing, 17(4):395–416, 2007. doi:https://doi.org/10.1007/s11222-007-9033-z.
https://doi.org/10.1007/s11222-007-9033-z
[35] Alexei Vázquez, Romualdo Pastor-Satorras, and Alessandro Vespignani. Large-scale topological and dynamical properties of the Internet. Physical Review E, 65(6):066130, 2002. arXiv:cond-mat/0112400. doi:https://doi.org/10.1103/PhysRevE.65.066130.
https://doi.org/10.1103/PhysRevE.65.066130
arXiv:cond-mat/0112400
[36] Stanley Wasserman and Katherine Faust. Social network analysis: Methods and applications. 1994. doi:https://doi.org/10.2307/3322457.
https://doi.org/10.2307/3322457
[37] Dorothea Wagner and Frank Wagner. Between min cut and graph bisection. In International Symposium on Mathematical Foundations of Computer Science, pages 744–750. Springer, 1993.
Cited by
[1] Simon Apers and Ronald de Wolf, “Quantum Speedup for Graph Sparsification, Cut Approximation and Laplacian Solving”, arXiv:1911.07306, (2019).
The above citations are from SAO/NASA ADS (last updated successfully 2023-07-04 02:00:04). The list may be incomplete as not all publishers provide suitable and complete citation data.
On Crossref’s cited-by service no data on citing works was found (last attempt 2023-07-04 02:00:02).
This Paper is published in Quantum under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. Copyright remains with the original copyright holders such as the authors or their institutions.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Automotive / EVs, Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- BlockOffsets. Modernizing Environmental Offset Ownership. Access Here.
- Source: https://quantum-journal.org/papers/q-2023-07-03-1046/
- :has
- :is
- :not
- $UP
- 1
- 10
- 11
- 12
- 13
- 14
- 15%
- 16
- 17
- 19
- 1985
- 1994
- 1996
- 1998
- 1999
- 20
- 2001
- 2005
- 2006
- 2008
- 2011
- 2012
- 2014
- 2015
- 2016
- 2017
- 2018
- 2019
- 2020
- 2021
- 22
- 23
- 24
- 25
- 26%
- 26th
- 27
- 28
- 30
- 30th
- 31
- 32
- 7
- 8
- 9
- a
- above
- ABSTRACT
- Academy
- access
- ACM
- ADvantage
- affiliations
- alex
- algorithms
- All
- allows
- American
- Amplification
- amsterdam
- an
- analysis
- and
- annual
- Application
- applications
- approximate
- ARE
- AS
- At
- Atsushi
- austin
- author
- authors
- average
- based
- BE
- becoming
- Benchmark
- Better
- between
- Beyond
- biology
- Blocks
- Bottom
- Brain
- Break
- Building
- but
- by
- CAN
- cannot
- capture
- challenging
- chan
- Chris
- class
- clustering
- CO
- comment
- Common
- Commons
- community
- comparison
- complete
- complex
- computations
- computer
- computer science
- computing
- Conference
- connected
- Connectivity
- constant
- Contagion
- contemporary
- context
- copyright
- counting
- Craig
- Cut
- CWI
- cycles
- Daniel
- Danielle
- data
- data mining
- David
- deeper
- Degree
- demonstrate
- Detection
- discovery
- discuss
- distributed
- distributed computing
- distribution
- dominant
- done
- dynamics
- e
- each
- Economic
- Edge
- efficient
- element
- Engineering
- Errors
- example
- expected
- Exploring
- extend
- fan
- faster
- fastest
- Fields
- Finally
- finance
- financial
- finding
- Focus
- For
- found
- Foundations
- from
- function
- General
- generating
- Genetics
- Gilles
- given
- graph
- graphs
- Group’s
- Grover
- harvard
- he
- help
- Hidden
- higher
- Highlighted
- highly
- holders
- However
- HTTPS
- IDO
- IEEE
- image
- impacted
- important
- in
- Including
- input
- insight
- institutions
- interesting
- International
- Internet
- into
- IT
- ITS
- JavaScript
- journal
- July
- knowledge
- known
- large
- large-scale
- Last
- Law
- leading
- learning
- Leave
- left
- less
- Level
- License
- like
- likely
- List
- listing
- Louis
- major
- make
- many
- Marcus
- maria
- mathematical
- mathematics
- Matrix
- max-width
- May..
- mcclean
- method
- methods
- Michael
- min
- Mining
- molecular
- Month
- more
- most
- mostly
- multiple
- Myths
- National
- Nature
- nearly
- Netherlands
- network
- networks
- Neuroscience
- no
- nodes
- obtain
- of
- often
- on
- ONE
- open
- or
- order
- organization
- original
- Other
- over
- pages
- Paper
- participate
- participation
- patterns
- performance
- performed
- Peter
- phase
- physical
- Physics
- plato
- Plato Data Intelligence
- PlatoData
- Popular
- power
- powerful
- presence
- present
- press
- Proceedings
- Progress
- properties
- provide
- provides
- providing
- published
- publisher
- publishers
- put
- quadratic
- Quantum
- quantum advantage
- quantum algorithms
- random
- recently
- Red
- references
- Regulation
- Relationships
- relative
- rely
- remains
- respect
- REST
- review
- Richard
- Risk
- Role
- RON
- royal
- Ryan
- s
- same
- Science
- SCIENCES
- scientific
- Search
- searching
- settings
- show
- siam
- Simon
- Simple
- Size
- small
- So
- Social
- social network
- Society
- Solving
- sophisticated
- Spectral
- speed
- Stability
- stanley
- statistical
- statistics
- steven
- straightforward
- strongly
- structure
- Study
- Studying
- subgraph
- subgraphs
- Successfully
- such
- suitable
- Sun
- Symposium
- systemic
- systemic risk
- Systems
- Testing
- that
- The
- the Netherlands
- their
- Them
- theoretical
- theory
- These
- this
- three
- Through
- time
- Title
- to
- together
- tom
- tool
- top
- Transactions
- transition
- tutorial
- two
- types
- uncover
- under
- underlying
- understand
- university
- updated
- URL
- use
- uses
- using
- usually
- various
- very
- volume
- von
- want
- was
- we
- web
- with
- Wolf
- works
- Workshops
- year
- zephyrnet